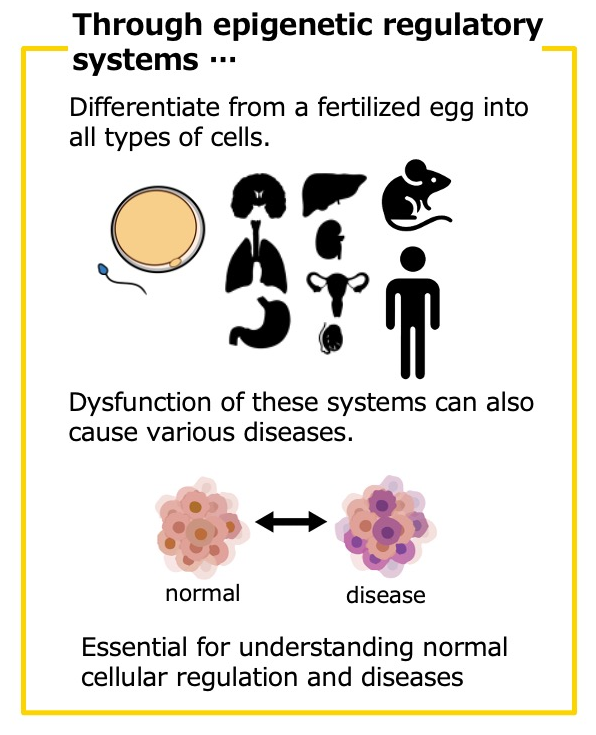
Various types of cells that form our body all carry the same DNA information. From this identical blueprint, diverse types and quantities of RNA are copied, which are used to produce the necessary proteins that determine the characteristics of each cell. The amount of RNA produced from each gene is usually profiled as gene expression levels and is associated with cell-type-specific information, linking the diverse phenotypes of normal cells and diseases. Epigenome is the set of chemical modifications to the DNA that facilitate such cell-type-specific gene expression from the "same blueprint" of DNA. DNA is wrapped around small proteins known as histones and compactly organized within the nucleus as chromatin. Histones undergo various chemical modifications, including methylation, acetylation, and ubiquitination, and DNA itself can also be modified with methylation.
These modifications regulate the chromatin structure, making it more open or closed, which either promotes or suppresses transcriptional activity across different genomic regions. These “epigenomic modifications”, which are maintained in a plastic manner with DNA, play a critical role in gene expression control, enabling a wide variety of phenotypes in both cell-type-specific and disease-specific contexts.
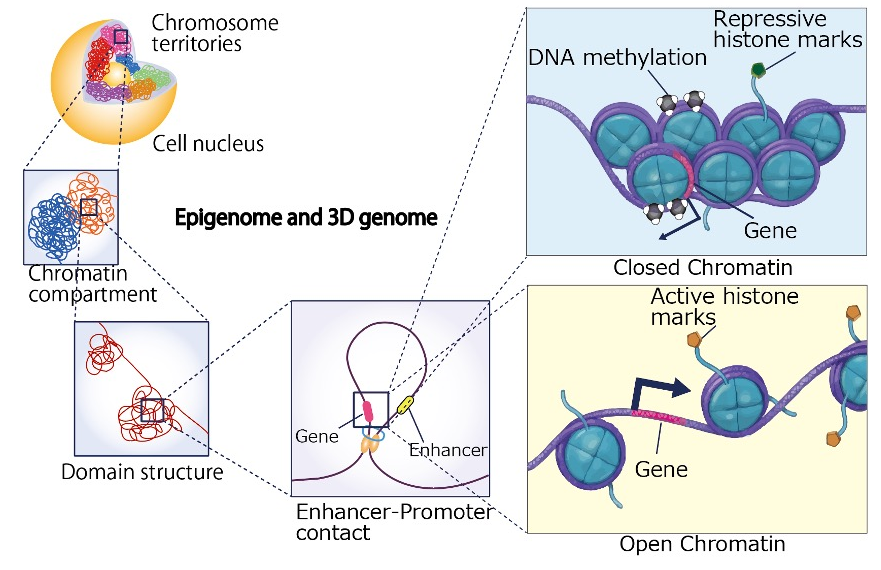
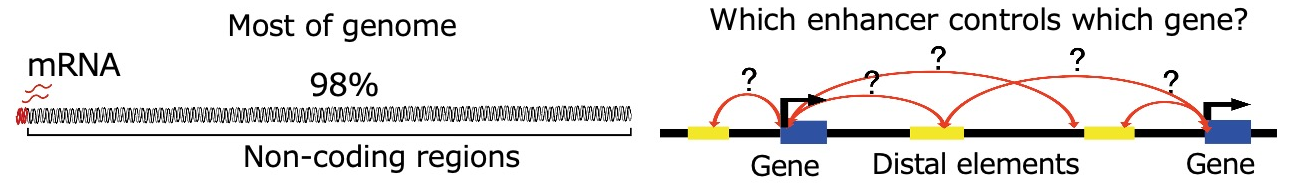
DNA stored within the nucleus as a chromatin structure consists of up to 3 billion base pairs in humans and mice. If the DNA from a single cell were stretched out, it would measure about 2 meters in length. However, it is not randomly packed inside the nucleus; instead, it is folded into an organized, three-dimensional structure (3D genome architecture). Importantly, the majority of our DNA consists of non-coding regions that do not encode proteins. Notably, these non-coding regions harbor various epigenomic modifications and contain numerous functional genomic elements, such as enhancers, that regulate transcription. However, even when we identify various epigenomic marks across these distal non-coding regions and predict active enhancer elements, it often remains unclear which genes they regulate. In this regards, 3D genome information, including DNA loop structures that spatially connect enhancers and promoters, provides crucial insights into how non-coding regions contribute to gene regulation.
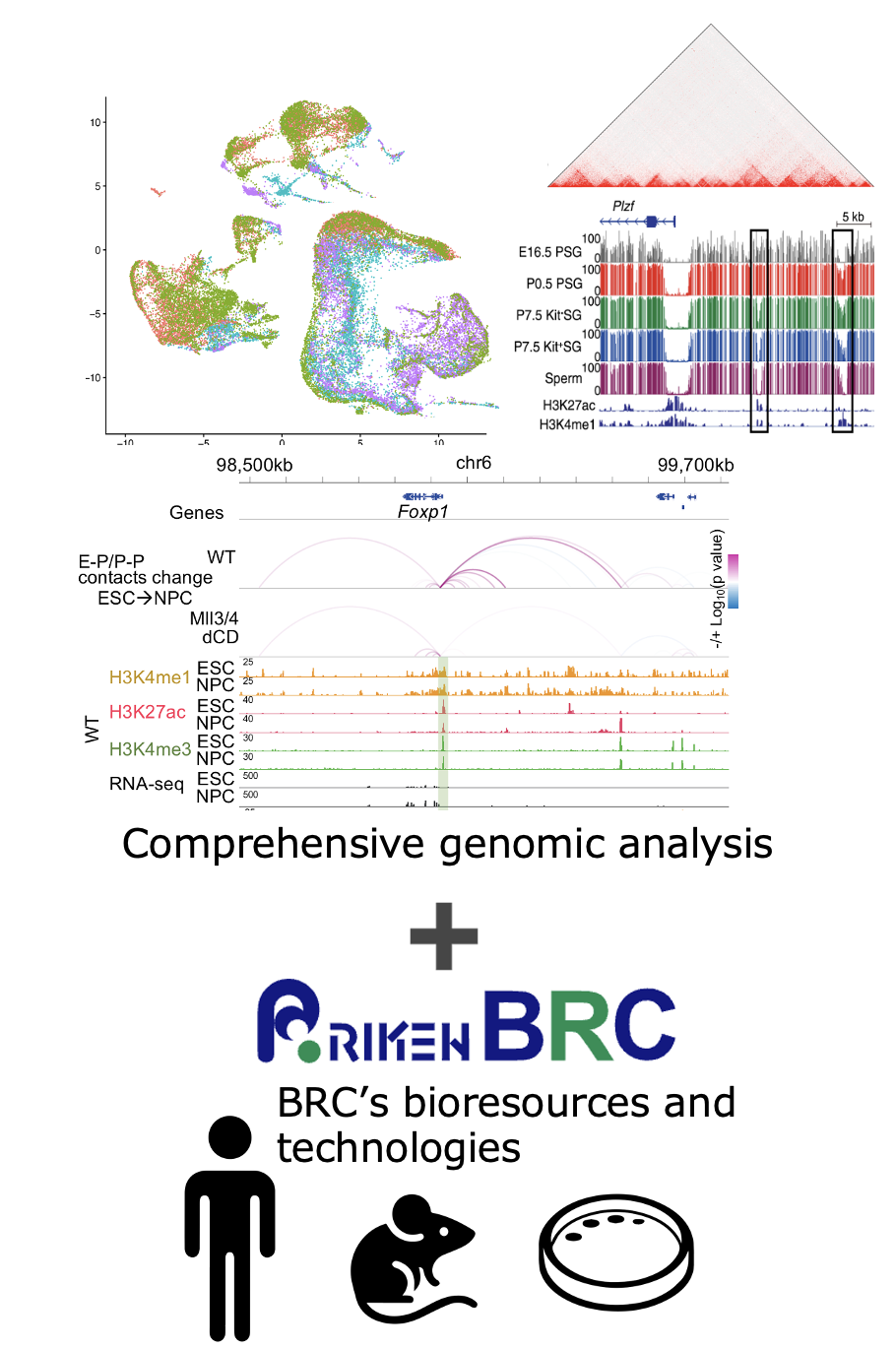
We have been conducting research on epigenetic regulatory mechanisms and 3D genome architecture as a transcriptional regulatory machinery using the latest analytical methods, focusing on mouse germ cells and ES cells (Publications).
At RIKEN BRC, we will introduce and develop state-of-the-art techniques of bulk and single-cell gene expression analysis, as well as epigenome and 3D genome analysis. By integrating these analytical approaches with artificial intelligence (AI) technologies, we aim to fully leverage the rich resources of model animals and iPS cell lines maintained at BRC to elucidate gene regulatory mechanisms underlying reproduction, development, and disease-specific processes.
Development of new analytical techniques to obtain gene expression, epigenome, and 3D genome information, along with the establishment of advanced bulk and single-cell analysis platforms.
Using these analytical technologies to elucidate gene expression regulatory mechanisms in reproduction, development, and diseases contexts.
Developing AI models to predict genome regulation and phenotypes using newly generated and accumulated epigenomic data.
Obtaining comprehensive genomic data corresponding to the diverse phenotypes of bioresources and the construction of a database.